Stop codon
In genetics, a stop codon or termination codon, also known as a nonsense codon, is a codon of ribonucleic acid (RNA) for which no associated tRNA (transfer RNA) is available and which therefore represents the end of a sequence of nucleotides that can be translated at ribosomes into the sequence of amino acids of a polypeptide.
A stop codon thus determines the end of a reading frame that allows the biosynthesis of a protein, and is thus a necessary condition for a coding nucleic acid segment. The base triplet of a stop codon on an mRNA (messenger RNA) leads to the termination of translation during protein biosynthesis in a cell and thus marks - similar to the dot at the end of a word sequence in a sentence - the end of the nucleotide sequence coding for a protein and thus its set of amino acids to be linked. The counterpart to the stop codon is the start codon at the beginning of this nucleotide sequence, where translation begins.
Three consecutive nucleic bases, a triplet, form the smallest meaning-bearing unit of the genetic code, called a codon. Each base triplet within an open reading frame codes for one of the proteinogenic amino acids that make up a polypeptide chain; a triplet of stop codons does not. These do not code for any amino acid, since there is no associated tRNA for these codons, but they define the end of a reading frame and thus, during translation, the end of the synthesis of a polypeptide chain for a protein.
In addition to the 61 base triplets of the standard genetic code that code for amino acids, there are three combinations of nucleic bases that can be used to terminate translation; these stop codons are located on the mRNA
UAGwith UAG = Uracil - Adenine - GuanineUGAwith UGA = uracil - guanine - adenineUAAwith UAA = Uracil - Adenine - Adenine
Under special conditions, the first two codons can also be interpreted in some organisms as coding for one amino acid each. The prerequisite for this is that a tRNA loaded with the respective amino acid is present, whose anticodon region binds to the codon on the mRNA. In order to distinguish the tRNA from the usual stop codon, additional circumstances are required, such as certain nucleotide sequences in the vicinity or special RNA structures such as hairpin formations. In some organisms, such conditions are such that the codon UAG can also be translated into the amino acid pyrrolysine, or the codon UGA into the amino acid selenocysteine. These two are therefore - in addition to the canonical twenty - also among the naturally occurring genetically encoded proteinogenic amino acids.
The role of nonsense mutations, in which a stop codon is created, was elucidated in the 1960s. In this context, the base triplet UAG was named amber after a member of the research group, Harris Bernstein. Subsequently, the triplet UAA was named ochre (ochre-colored) and the triplet UGA was named opal (opal-colored). These names are an allusion, since the colors have physically and chemically nothing to do with the base triplet.
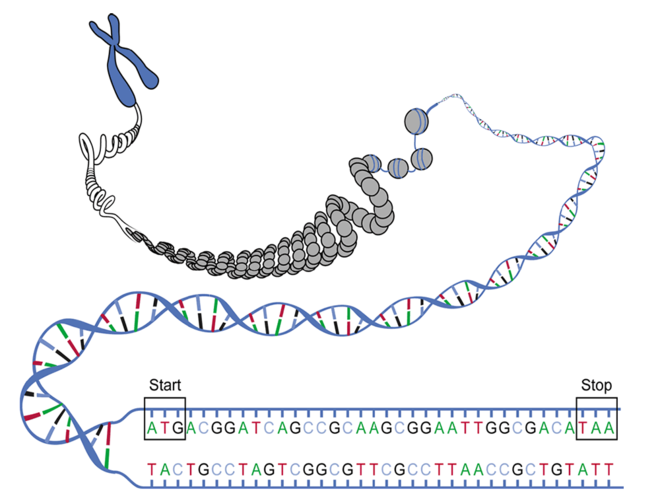
Schematic representation of a DNA double strand whose gene region is unwound for transcription and separated into two single-stranded segments. On the non-matrix DNA strand shown above, a base sequence is located between "start" and "stop", complementary to that of the matrix strand (bottom). At this codogenic DNA strand as a template, an RNA strand is built up as a transcript with the help of RNA polymerase, which becomes the messenger RNA. In the example, the nucleotide sequence of this mRNA can then contain the following codons as a series of base triplets in the reading frame beginning with the start codon: AUGACGGAUCAGCCGCAAGCGGAAUUGGCGACAUAA. The last one is the stop codon.
Mutants
Bacterial mutants whose mRNA contains the codon UAG resulting from point mutation are also called amber mutants. A compensatory mutation in a tRNA gene can enable the protein-synthesizing system here to interpret this codon as a sense codon. With the corresponding tRNA gene in the genome of an organism, the codon UAG can then be translated into a (proteinogenic) amino acid.
If point mutations lead to a stop codon (so-called nonsense mutation), this usually results in a shortened protein (truncating mutation), provided the mutation is not within an intron that is dropped during splicing. For the conversion of the start codon AUG into a stop codon, two hitting point mutations would be required in the case of UAG, and three in the case of UGA and UAA.
Search within the encyclopedia