Coronaviridae
![]()
This article is about the family of coronaviruses. For the current disease incidence of one of these viruses, see COVID-19, for the spread of the disease COVID-19 pandemic and its cause SARS-CoV-2.
Coronaviridae is a family of viruses within the order Nidovirales. The viruses within the family are colloquially called "coronaviruses" and belong to the RNA viruses with the largest genomes.
The first coronaviruses were described as early as the mid-1960s. The British virologist June Almeida is considered to be the discoverer, who succeeded in taking a picture using an electron microscope in 1966. The roughly spherical viruses in the electron microscope image are conspicuous for their ring of petal-like projections, which are reminiscent of a solar corona and which gave them their name.
Representatives of this virus family cause very different diseases in all four classes of terrestrial vertebrates (mammals, birds, reptiles, amphibians). They are genetically highly variable and can thus sometimes infect several species of hosts. In humans, seven types of coronaviruses are important as pathogens of mild respiratory infections (cold / flu) up to the so-called Severe acute respiratory syndrome (SARS).
Among human coronaviruses, the following have become particularly well known:
- SARS-CoV[-1] (severe acute respiratory syndrome coronavirus [1])
- MERS-CoV (Middle East respiratory syndrome coronavirus)
- SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2)
They were or are the triggers of the 2002/2003 SARS pandemic, the MERS epidemic (from 2012) and the COVID 19 pandemic (from 2019).
Note: For a clearer and more meaningful distinction between the coronaviruses SARS-CoV and SARS-CoV-2, SARS-CoV is sometimes also referred to as SARS-CoV-1.
Features
Appearance
The 60 to 160 nm large virus particles (virions) have a viral envelope in which several different membrane proteins are embedded. The characteristic appearance of coronaviruses (Latin corona 'wreath, crown') is due to many club-shaped structures on the surface, called peplomers, which project outwards by about 20 nm. They consist of portions of the large glycosylated S protein (spikes protein, 180 to 220 kDa), which here forms a membrane-anchored trimer. These parts carry both (S1) the receptor-binding domain (RBD), with which the virus can dock onto a cell, and (S2) a subunit which, as a fusion protein (FP), brings about the fusion of the viral envelope and the cell membrane.
The smaller envelope protein (E protein, 9 to 12 kDa) is present in smaller quantities on the outside. Only in HCoV-OC43 (human coronavirus OC43) and the coronaviruses of group 2 (genus Betacoronavirus) is the haemagglutin esterase protein (HE protein, 65 kDa) also found. In contrast, the M protein (matrix protein, 23 to 35 kDa), which is also anchored in the membrane envelope, is directed inwards and is a matrix protein on the inside of the viral envelope.
Inside the envelope is a presumably icosahedral capsid containing a helical nucleoprotein complex. This consists of the nucleoprotein N (50 to 60 kDa) complexed with the strand of a single-stranded RNA of positive polarity. Certain amino acid residues of the N protein interact with the matrix protein M so that the capsid is associated with the membrane interior.
Genome
The single-stranded RNA genome of coronaviruses is approximately 27,600 to 31,000 nucleotides (nt) long, making coronaviruses the longest genomes of all known RNA viruses.
At the 5' end there is a 5' cap structure and a non-coding region (untranslated region, UTR) of about 200 to 400 nt, which contains a 65 to 98 nt short, so-called leader sequence. At the 3' end, another UTR of 200 to 500 nt is added, which ends in a poly(A) tail. The coronavirus genome contains 6 to 14 open reading frames (ORFs), of which the two largest (the genes for the non-structural proteins NSP-1a and 1b) are located near the 5' end and overlap somewhat with different reading frames. The overlap site forms a hairpin structure that allows a reading frame jump in translation at ribosomes in 20 to 30% of the reading runs, leading to the synthesis of lower amounts of NSP-1b.
In addition to replicating their genome, viruses synthesize 4-9 mRNA molecules (depending on the virus species) whose 5' and 3' ends are identical to those of the genome. These "nested" mRNAs are also referred to as "nested set of mRNAs" and have contributed to the naming of the superordinate viral order, Nidovirales (from Latin nidus 'nest').
In contrast to the usually high error rate of the RNA polymerase of other RNA viruses, which leads to a restriction of the genome length to about 10,000 nucleotides, in coronaviruses a relatively high genetic stability (conservation) is achieved, among other things, by a 3'-5' exoribonuclease function of the protein NSP-14. Presumably, this proofreading mechanism causes the antiviral agent ribavirin to be ineffective in COVID-19 (SARS-CoV-2).
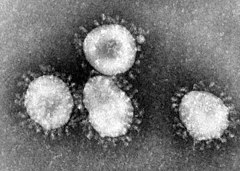
Electron micrograph of coronaviruses
Occurrence and distribution
Infectious bronchitis in poultry caused by infectious bronchitis virus (IBV, species Avian coronavirus), a gammacoronavirus of the subfamily Orthocoronavirinae, was studied as early as 1932. At that time, investigations focused on the disease process. The appearance and genetic relationships of the virus were unknown and the name "coronaviruses" did not yet exist.
The first named coronaviruses were described in the mid-1960s. The first specimen discovered was the later lost human coronavirus B814 (unclassified). Coronaviruses are genetically highly variable; individual species from the family Coronaviridae can also infect several species of hosts by overcoming the species barrier, thus causing zoonoses.
Overcoming the species barrier has resulted in human infections with, among others, SARS coronavirus (SARS-CoV, sometimes referred to as SARS-CoV-1) - the causative agent of the 2002/2003 SARS pandemic - and Middle East respiratory syndrome coronavirus (MERS-CoV), a new virus that emerged in 2012.
The COVID-19 pandemic that originated in the Chinese city of Wuhan in early 2020 is attributed to a previously unknown coronavirus that has been named SARS-CoV-2.
Questions and Answers
Q: What is Coronaviridae?
A: Coronaviridae is a family of enveloped, positive-sense, single-stranded RNA viruses.
Q: What is the length of the viral genome of Coronaviridae?
A: The viral genome of Coronaviridae is 26-32 kilobases in length.
Q: How do the particles of Coronaviridae look like?
A: The particles of Coronaviridae have large (~20 nm), club- or petal-shaped spikes on the surface known as the "peplomers."
Q: What is the appearance of the particles of Coronaviridae in electron micrographs?
A: In electron micrographs, the particles of Coronaviridae resemble the solar corona.
Q: What is the significance of the "peplomers" on the surface of the particles of Coronaviridae?
A: The "peplomers" on the surface of the particles of Coronaviridae are significant as they aid in attachment to the host cell and play a role in the virulence of the virus.
Q: Is Coronaviridae a DNA or RNA virus?
A: Coronaviridae is an RNA virus.
Q: What is the size of the spikes on the surface of the particles of Coronaviridae?
A: The spikes on the surface of the particles of Coronaviridae are approximately 20 nm in size.
Search within the encyclopedia